This function creates an average metagene for gene clusters.
createMetagenes( gobject, expression_values = c("normalized", "scaled", "custom"), gene_clusters, name = "metagene", return_gobject = TRUE )
Arguments
| gobject | Giotto object |
|---|---|
| expression_values | expression values to use |
| gene_clusters | numerical vector with genes as names |
| name | name of the metagene results |
| return_gobject | return giotto object |
Value
giotto object
Details
An example for the 'gene_clusters' could be like this: cluster_vector = c(1, 1, 2, 2); names(cluster_vector) = c('geneA', 'geneB', 'geneC', 'geneD')
Examples
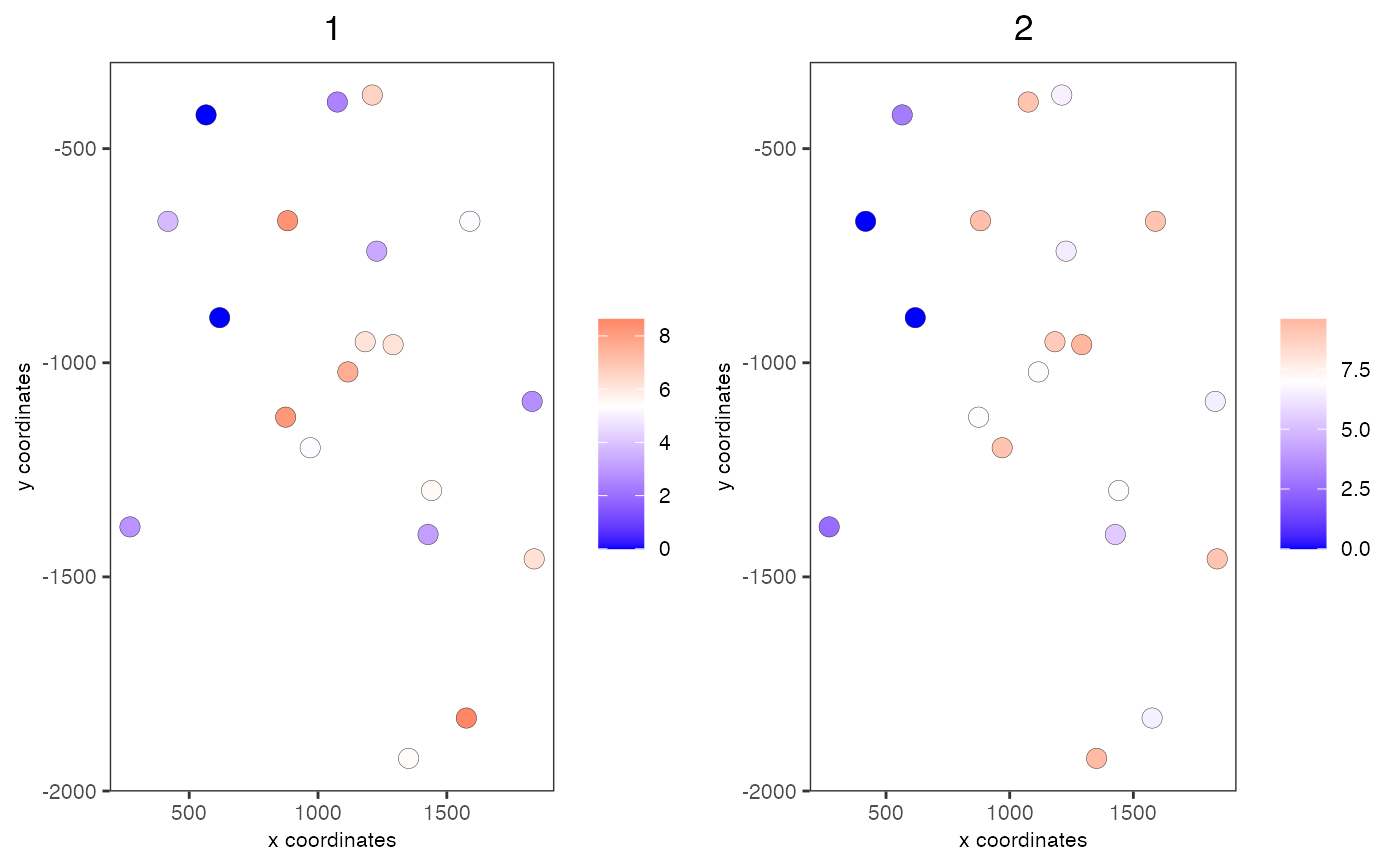
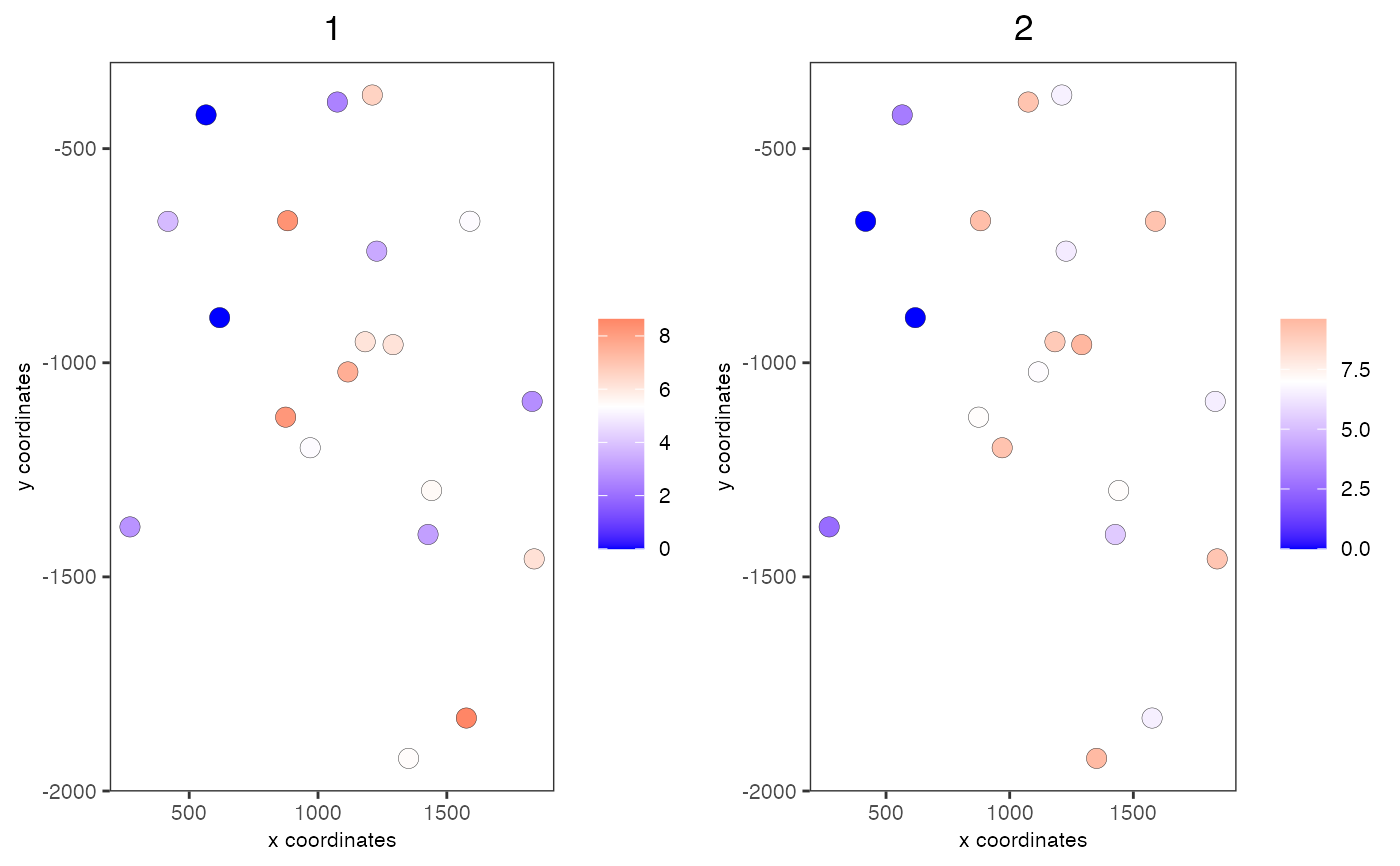
data(mini_giotto_single_cell) # get all genes all_genes = slot(mini_giotto_single_cell, 'gene_ID') # create 2 metagenes from the first 6 genes cluster_vector = c(1, 1, 1, 2, 2, 2) # 2 groups names(cluster_vector) = all_genes[1:6] mini_giotto_single_cell = createMetagenes(mini_giotto_single_cell, gene_clusters = cluster_vector, name = 'cluster_metagene')#> #> cluster_metagene has already been used, will be overwritten# show metagene expression spatCellPlot(mini_giotto_single_cell, spat_enr_names = 'cluster_metagene', cell_annotation_values = c('1', '2'), point_size = 3.5, cow_n_col = 2)