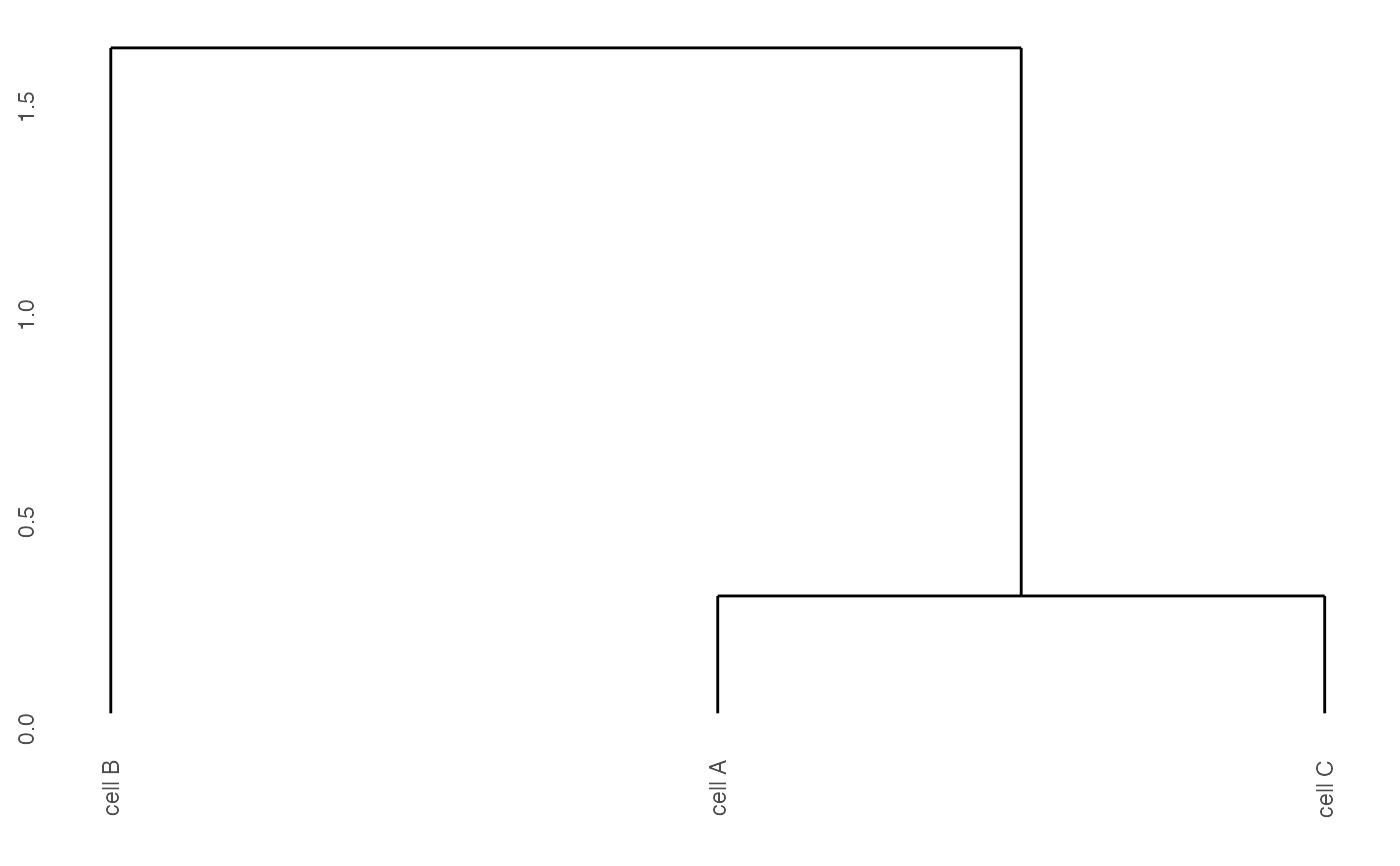
Creates dendrogram for selected clusters.
showClusterDendrogram( gobject, expression_values = c("normalized", "scaled", "custom"), cluster_column, cor = c("pearson", "spearman"), distance = "ward.D", h = NULL, h_color = "red", rotate = FALSE, show_plot = NA, return_plot = NA, save_plot = NA, save_param = list(), default_save_name = "showClusterDendrogram", ... )
Arguments
| gobject | giotto object |
|---|---|
| expression_values | expression values to use |
| cluster_column | name of column to use for clusters |
| cor | correlation score to calculate distance |
| distance | distance method to use for hierarchical clustering |
| h | height of horizontal lines to plot |
| h_color | color of horizontal lines |
| rotate | rotate dendrogram 90 degrees |
| show_plot | show plot |
| return_plot | return ggplot object |
| save_plot | directly save the plot [boolean] |
| save_param | list of saving parameters, see |
| default_save_name | default save name for saving, don't change, change save_name in save_param |
| ... | additional parameters for ggdendrogram() |
Value
ggplot
Details
Expression correlation dendrogram for selected clusters.
Examples
data(mini_giotto_single_cell) # cell metadata cell_metadata = pDataDT(mini_giotto_single_cell) # create heatmap showClusterDendrogram(mini_giotto_single_cell, cluster_column = 'cell_types')