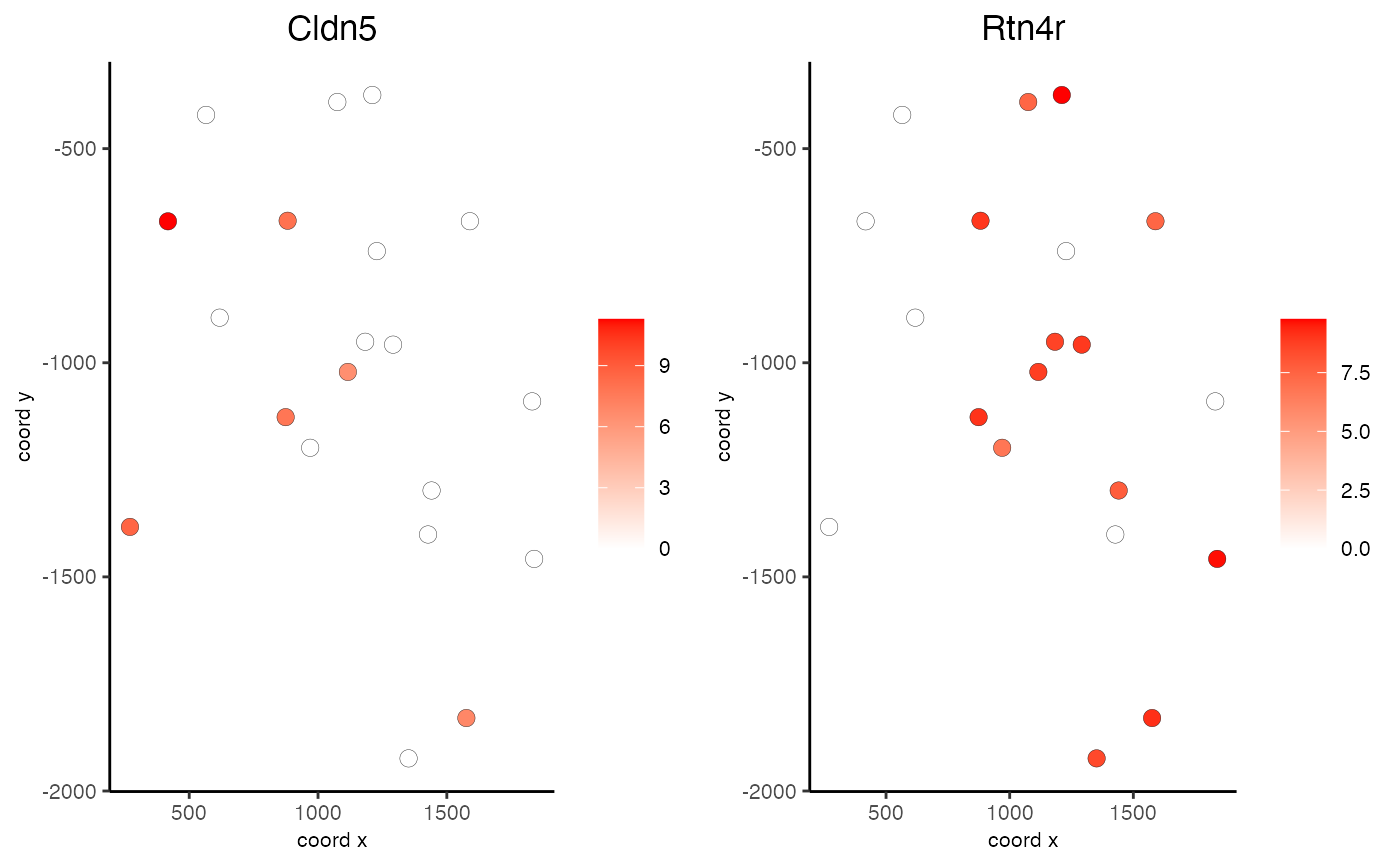
Visualize cells and gene expression according to spatial coordinates
spatGenePlot(...)
Arguments
| ... |
Arguments passed on to spatGenePlot2D
gobjectgiotto object show_imageshow a tissue background image gimagea giotto image image_namename of a giotto image sdimxx-axis dimension name (default = 'sdimx') sdimyy-axis dimension name (default = 'sdimy') expression_valuesgene expression values to use genesgenes to show cell_color_gradientvector with 3 colors for numeric data gradient_midpointmidpoint for color gradient gradient_limitsvector with lower and upper limits show_networkshow underlying spatial network network_colorcolor of spatial network spatial_network_namename of spatial network to use edge_alphaalpha of edge show_gridshow spatial grid grid_colorcolor of spatial grid spatial_grid_namename of spatial grid to use midpointexpression midpoint scale_alpha_with_expressionscale expression with ggplot alpha parameter point_shapeshape of points (border, no_border or voronoi) point_sizesize of point (cell) point_alphatransparancy of points point_border_colcolor of border around points point_border_strokestroke size of border around points cow_n_colcowplot param: how many columns cow_rel_hcowplot param: relative height cow_rel_wcowplot param: relative width cow_aligncowplot param: how to align show_legendshow legend legend_textsize of legend text background_colorcolor of plot background vor_border_colorborder colorr for voronoi plot vor_max_radiusmaximum radius for voronoi 'cells' vor_alphatransparancy of voronoi 'cells' axis_textsize of axis text axis_titlesize of axis title show_plotshow plots return_plotreturn ggplot object save_plotdirectly save the plot [boolean] save_paramlist of saving parameters, see showSaveParameters default_save_namedefault save name for saving, don't change, change save_name in save_param |
Value
ggplot
Details
Description of parameters.
See also
Examples